Tracking Translation of Human Genome Discoveries into Prevention and Control of Common Chronic Diseases: The Action is in Cancer!
Posted on by Genomics seems to be everywhere these days. From the Human Genome Project to the Precision Medicine Initiative and from the Cancer Moonshot to breakthroughs in genome editing, we are overwhelmed with information about genomics. The hope and hype of discoveries are intermixed daily in published scientific articles and media coverage of how they might improve our health. However, regardless of the hype, most of the field is still in its infancy. Although ongoing progress can be measured by the volume of published papers, the real test is how these discoveries will be translated into practice and what the health benefits will be, if any. In our opinion, this is still a “road less traveled.”
Genomics seems to be everywhere these days. From the Human Genome Project to the Precision Medicine Initiative and from the Cancer Moonshot to breakthroughs in genome editing, we are overwhelmed with information about genomics. The hope and hype of discoveries are intermixed daily in published scientific articles and media coverage of how they might improve our health. However, regardless of the hype, most of the field is still in its infancy. Although ongoing progress can be measured by the volume of published papers, the real test is how these discoveries will be translated into practice and what the health benefits will be, if any. In our opinion, this is still a “road less traveled.”
Chronic diseases are a major public health problem and account for a large burden of morbidity, mortality, and disability in the United States and around the world. Chronic diseases are responsible for 7 of 10 deaths each year, and treating people with chronic diseases accounts for 86% of our nation’s health care costs. Can genomics help in the prevention and control of chronic disease? Are there current guidelines and recommendations for genomics in chronic diseases?
We decided to take a look at the CDC Public Health Genomics Knowledge Base (PHGKB) to track progress in genomics translation for top common chronic diseases. PHGKB is a continuously updated online searchable knowledge base of the evolving landscape of genomic applications in health care and disease prevention. The knowledge base is geared to researchers, health policy makers, and practitioners. Details on PHGKB are available in a previous blog posting and a recent scientific article.
We searched PHGKB for published translational studies, guidelines and recommendations and implementation tools and studies for the top common chronic diseases: cancer, heart disease/stroke, diabetes, arthritis and Alzheimer’s disease. As shown in the accompanying graphs, we found that cancer accounted for most of the “action” for potential genomics translation into healthcare and disease prevention.
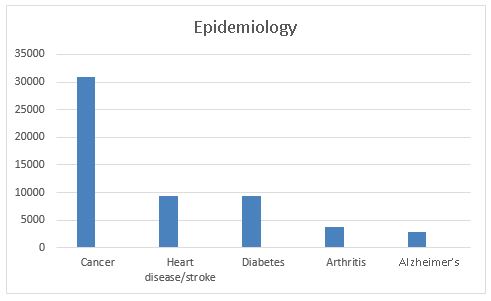
Let’s elaborate. The first graph shows the number of human genomic epidemiologic studies published since 2001 for these diseases. Epidemiologic studies are crucial in genomics not only to document associations between genes and diseases but to assess the relationship in terms of risks and interactions among genes and environmental factors. This is a necessary first step of genomic translation. There were almost 31,000 articles in cancer, more than the number of articles for all the other diseases combined. We did not review specific articles for this analysis. PHGKB users can more fully examine these results looking at specific genes and diseases, and retrieving articles, systematic reviews, and meta-analyses.
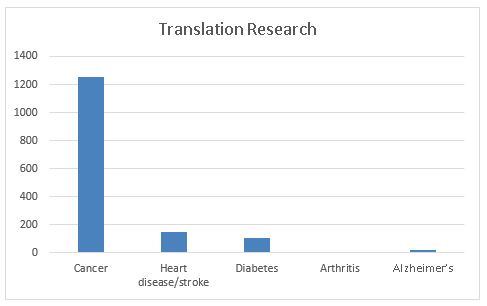
The second graph documents the volume of “translational research” that is “beyond bench to bedside.” This research attempts to evaluate the health benefits of genomic tests and targeted interventions and how they can be delivered, and documents their effectiveness and outcomes in real world practice. We have shown previously that this research constitutes about 1% or less of all published genomic studies. The graph also shows the large dominance of cancer in this type of research (more than 1200 articles).
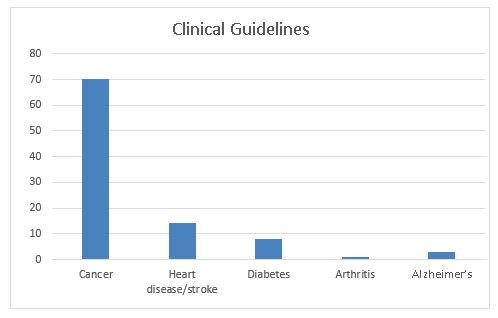
The third graph shows the number of published professional guidelines and recommendations for the use of genomics in practice. Here again, we see the large excess in cancer (70 guidelines, compared to 10 or less for each of the other conditions). The guidelines cover the spectrum of targeted prevention, treatment, and prognosis based on genomic markers.
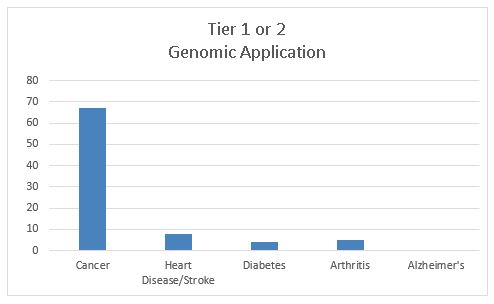
The fourth graph shows the most important indicator for success: the existence of genomic tests and other applications that have been recommended for clinical practice. We use a systematic process and label these applications using several tiers. For “tier 1” applications (tests that have been recommended for use by evidence-based panels), the cancer field has 34 entries compared to only 2 for all other diseases combined. In addition, genomic tests that have evidence of clinical validity but not enough evidence for utility, or “tier 2” applications (tests with adequate information on clinical validity but no evidence-based guidelines for their use), also predominate for cancer. Thirty-three tier 2 applications exist for cancer genomics compared to 15 for all other diseases combined. Readers can peruse the list of these applications as they pertain to a whole range of practice including prevention, screening, and treatment.
Perhaps it is not surprising to document that the current clinical and public health action in genomics is in cancer. While inherited susceptibility is common to all chronic diseases, cancer has the main distinction that it directly involves gene mutations that lead to cell changes and tumor development. Genetic changes can be used for early diagnosis and to target preventive and curative interventions. Several applications of cancer genomic biomarkers are currently recommended in practice covering the spectrum of targeted treatment, early detection, and prevention. With ongoing initiatives such as the Precision Medicine Initiative and the Cancer Moonshot, clinical and public health action in cancer genomics is likely to mature more quickly than other chronic diseases. It remains to be seen, however, to what extent the field of genomics can help reduce the burden of cancer in the United States and around the world.
Number of publications for selected common chronic diseases, by type of publication.
For details on types of publication, please see text and detailed descriptions on the Public Health Genomics Knowledge Base website https://phgkb.cdc.gov/GAPPKB/phgHome.do?action=home
Search of the database was conducted on October 11, 2016
Posted on by