From the Genome to the Exposome: Mapping Causal Associations Between Environmental Factors and Population Health
Posted on by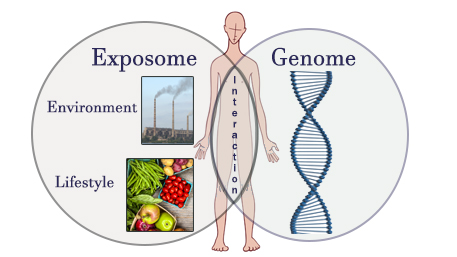 Environmental factors such as lifestyle, diet, and exposure to toxins and chemical agents, can play an important role in our health. Complementary to the “genome,” which is the complete set of an individual’s genetic information, the “exposome” represents an individual’s complete set of environmental exposures throughout their lifetime. Coined in 2005, the “exposome” captures the variable and dynamic environmental exposures from the prenatal period onwards. Factors studied in the exposome extend from social differences and inequalities to behavioral factors such as tobacco smoking and physical activity. Our genome and exposome interact throughout our lives in causing various diseases.
Environmental factors such as lifestyle, diet, and exposure to toxins and chemical agents, can play an important role in our health. Complementary to the “genome,” which is the complete set of an individual’s genetic information, the “exposome” represents an individual’s complete set of environmental exposures throughout their lifetime. Coined in 2005, the “exposome” captures the variable and dynamic environmental exposures from the prenatal period onwards. Factors studied in the exposome extend from social differences and inequalities to behavioral factors such as tobacco smoking and physical activity. Our genome and exposome interact throughout our lives in causing various diseases.
Probing the effects of the highly dynamic exposome on population health is challenging. One approach is the environment-wide association study (EWAS). Inspired by the concept of genome-wide association study (GWAS), which scans the entire genome to identify genes associated with a particular trait or disease, EWAS takes an untargeted, agnostic, and exploratory approach to identifying available environmental factors associated with a particular trait or disease. By broadening the search space to a wide array of environmental traits, the EWAS approach represents a promising technique for identifying novel factors in a systematic fashion, an approach that may aid in the development of disease prevention and intervention tactics.
As an example, a recent investigation published in JAMA Psychiatry conducted an exposome-wide study and Mendelian randomization (MR) analysis to investigate a range of nongenetic factors associated with psychotic experiences. MR is a method that leverages genetic differences to reduce bias from confounding variables and infer causal relationships. Investigators analyzed data from 155,247 participants in the UK Biobank, a large cohort study from the United Kingdom. First, they performed an exposome-wide study to identify environmental correlates with psychotic experiences. For variables that remained statistically significant when tested in a multivariable model, the authors conducted MR studies to probe for evidence of causality. They identified 148 environmental exposures, including traumatic experiences, hearing difficulties, and neighborhood, social, and economic deprivation. They also identified factors that have been previously unexplored in association with psychotic experience, such as nitrogen dioxide air pollution. This study highlights the potential of coupling systematic exposome-wide analyses with genetically-informed approaches to test for causality as a means for unraveling novel modifiable risk factors associated with disease.
Limitations of the study include the use of stringent multiple-testing correction, which may increase the chance for missing an association in the exposome-wide analysis. The MR analyses are also limited by weak instrument bias and genetic variants possibly influencing multiple phenotypes.
Environmental exposures have been shown to influence the incidence and severity of many chronic conditions. Discovering environmental correlates of disease by using exposome-wide analyses coupled with genetically anchored tests, such as MR, to help determine causality can identify modifiable exposures and behaviors that may influence disease risk. The Human Genome Project unlocked our DNA blueprint and allowed for genetics to be an important tool for investigating causality of the environment in EWAS. Advancing these research efforts is important for strengthening the next generation public health strategies and interventions, driving us forward toward precision public health.
Posted on by


