Tracking the Scientific Literature on SARS-CoV-2 Variants Using the COVID-19 Genomics and Precision Health Knowledge Base
Posted on by
The first reports of SARS-CoV-2, the highly infectious virus causing COVID-19, swept across the globe in December 2019, prompting a burst of scientific activity. The rate of research and discovery intensified as the pandemic grew, resulting in a flood of publications in journals and on preprint servers around the world. More recently, SARS-CoV-2 variants have become a major focus of SARS-CoV-2 research in basic, clinical, and public health sciences.
CDC’s Office of Genomics and Precision Public Health established the COVID-19 Genomics and Precision Health (COVID-19 GPH) database to capture publications that reflect the influence of two broad emerging technologies: genomics (pathogen and human), and precision health (machine learning, artificial intelligence, and predictive analytics). Together, these fields are the leading edge of precision public health in COVID-19 and beyond. Data are continuously updated from PubMed, the NIH iSearch COVID-19 Portfolio, LitCovid, and media sources using an automatic retrieval and text mining strategy3 and manual curation by CDC staff.
To examine trends in published research on SARS-CoV-2 variants, we use COVID-19 GPH built-in search tools, restricting our analysis to publications containing the words “variant” or “variants.” The number of COVID-19 publications are displayed graphically by month and year.
From December 2020 to January 2021, the number of preprints and journal publications on variants doubled from 106 to 213 (Figure 1). By March 2021, more than 300 new publications on variants were appearing each month. Preprints accounted for 37% of the total publications in both 2020 and 2021.
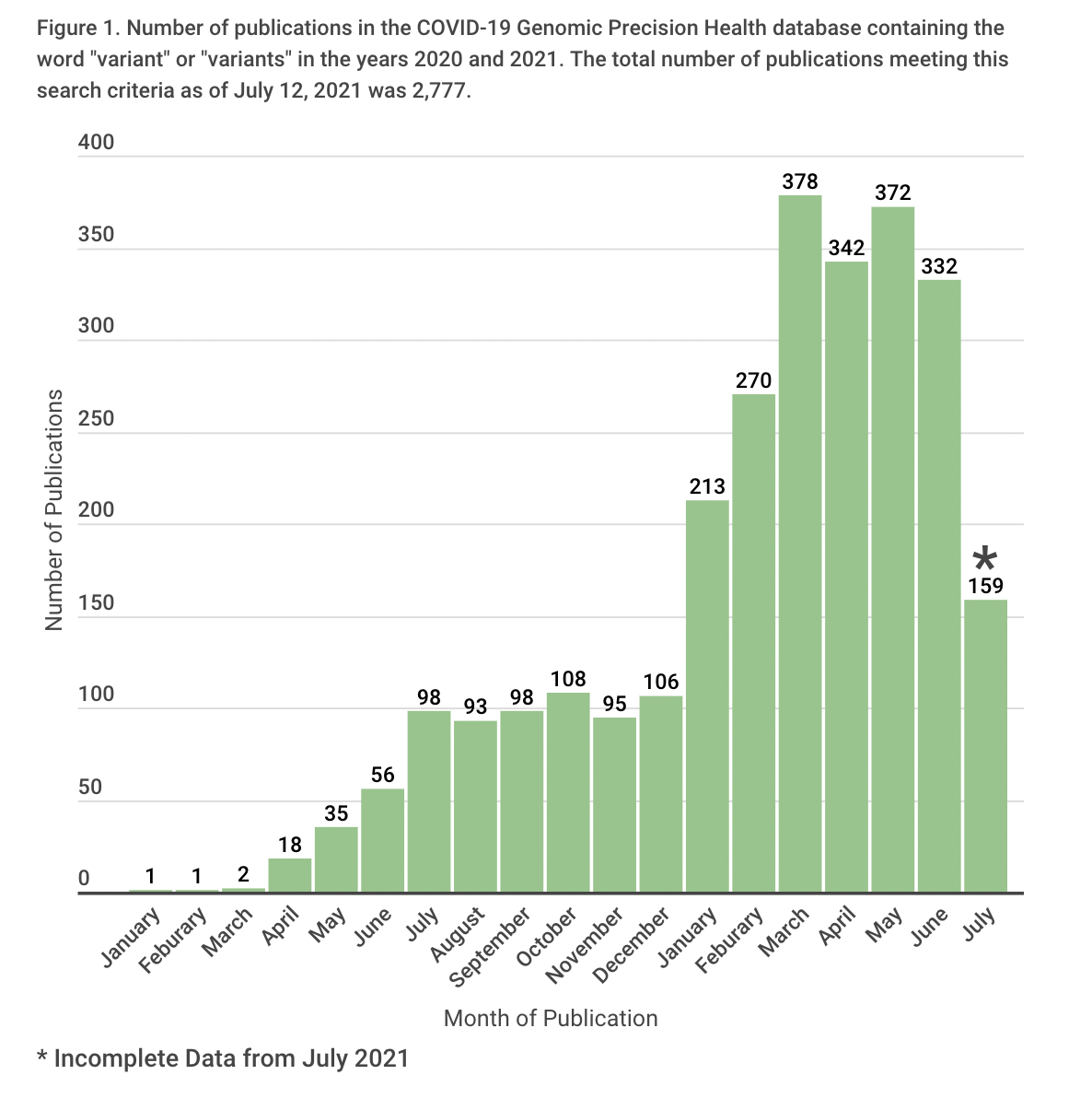
We also examined categories of publications based on eight indexing terms assigned by LitCovid and three more terms (vaccine, variant, health equity) assigned by COVID-GPH using text mining scripts. Records can be indexed by more than one term. We limited our analysis to records identified by the variant indexing term; frequencies of the other terms are displayed in Figure 2. Most publications on variants focused on mechanism (e.g., effects of mutations on binding to host receptors) or on potential clinical applications (e.g., diagnosis, treatment, or vaccines). Only a small fraction addressed public health topics, such as surveillance or forecasting, prevention, or health equity.
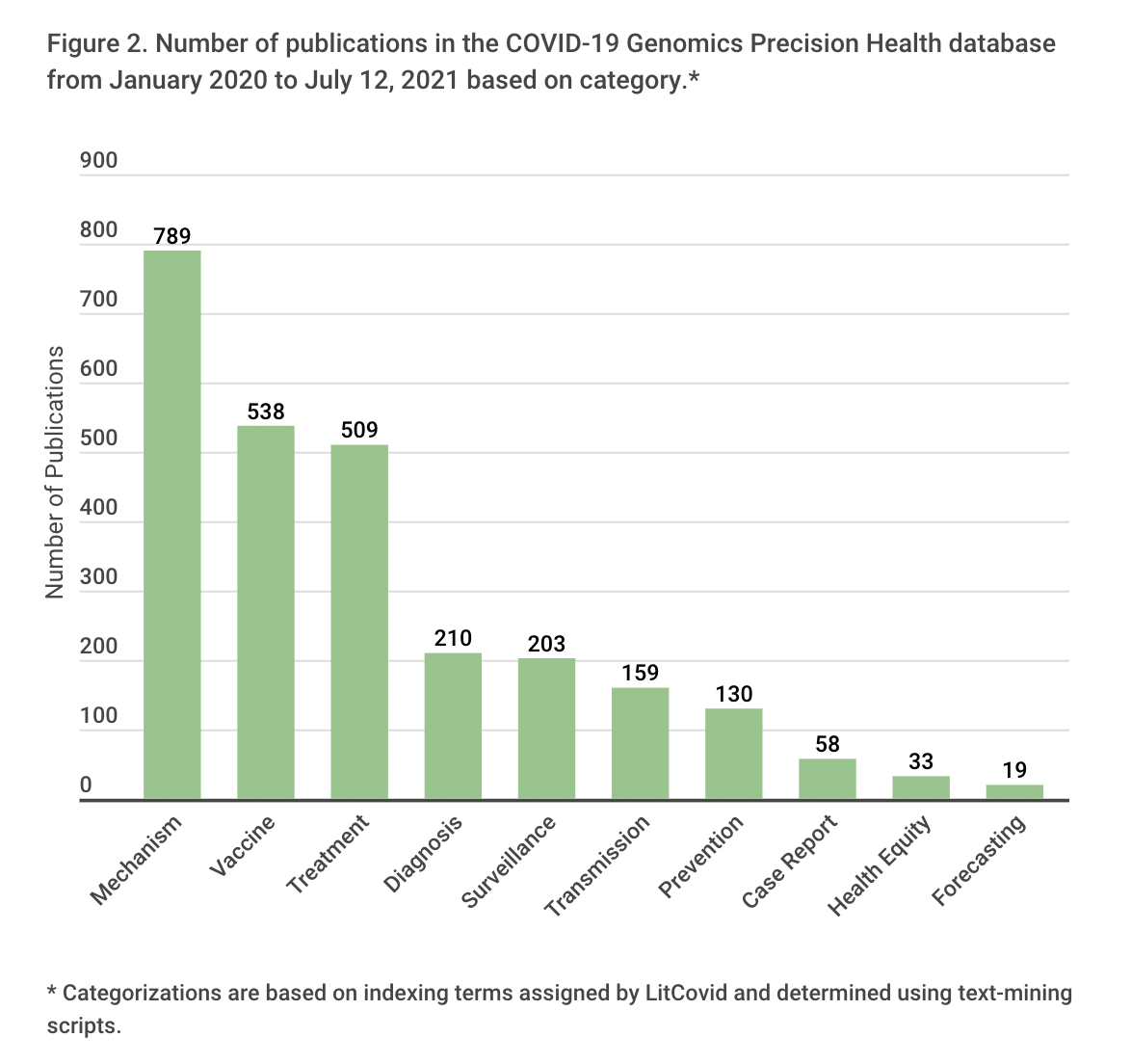
In summary, the COVID19- GPH database clearly shows an upward trajectory in the number and scope of publications on SARS-CoV-2 variants in the last few months, reflecting the response of the scientific community to assessing their impact on COVID-19 occurrence, transmission, outcomes, prevention, and treatment. CDC is conducting genomic surveillance of SARS-CoV-2 to detect emerging variants, study their characteristics, and monitor their frequencies over time. CDC’s Office of Advanced Molecular Detection has developed the COVID-19 Genomic Epidemiology Toolkit to help epidemiologists integrate SARS-CoV-2 genomics into state and national efforts for precision public health surveillance, outbreak investigation, and control of COVID-19.
We encourage our readers to check out COVID-19-GPH regularly as a timely and updated source of information and publications on COVID-19 and precision public health.
Posted on by


